Participant 1
- According to FDA, we are encouraged to use codelists on more variables than the ones in the SEND IG that are referring to “official” NCI codelists. What criteria should we use to choose these extra variables? At first sight, there are some evident choices like f.ex. lbcat or lbscat. But for other variables which end up having a short number of distinct values in the data, and for which you do not have a clear indication beforehand of the “potential” values: should these be chosen to use custom codelist? [2]
- FDA also encourages us not to merge codelists across many variables. The typical example is UNIT, being used across many variables in the SEND IG. So if we defined a “BGUNIT” codelist for the variable BGORRESU: is it OK to use the same codelist for the BGSTRESU variable? Or should we use one codelist per variable (instead of one codelist for the 2 variables)? Or should this choice be dependent on the Origin attribute of the variables [4]
- Should codelists contain the range of possible values for a specific variable for a specific study? Or only the actual values found in the actual data? We believe the first option is the right one so: should we explain in the nSDRG the reason for the inclusion of values not found in the actual data? [3]
Participant 2
- For define file code list, the question I have is how FDA is going to use this code list and terms included in the define file. Will FDA load this to its data warehouse for each study and use this to compare against dataset? [1]
- Three possible ways for including code list terms in define file, which is appropriate? [3]:
1. All available terms used in the code list, including CDISC published terms and user define terms (for extensible code list).
2. Only terms used in the study, including CDISC published terms and user define terms
3. Only user defined terms for the study
Participant 3
- Should the define file include all available terms in the codelist (CDISC and user defined terms) or only the terms used in the study? Ideally, it will be easier to implement if it is consistent across all codelists, not dependent on individual ones. [3]
- Should user defined codelists (nonCDISC codelists) be included in the define file?
- Should user defined codes use the CDISC code if it exists in a future CT package, or should it use a nonstandard code? It is understood that this would still be an extended term to the CT package used to generate the dataset. [8]
Participant 4
is what should / should not be included in the comments for the Define.xml?
Note from J. Feldmann: Codelists don't have comments - need clarification on this question.
Participant 5
here is some of the feedback we received from the FDA test submission reviewer regarding the use of code lists in the define.xml file. It would be great to get some additional clarity and consensus between industry and the agency. As background, our submission included 4 variables that leveraged the UNITS code list and the code list only contained the four units included in the study’s data.[4]
Some Codelists are merged across many variables
• Codelist should describe variable collection or derivation. Therefore, Codelist is expected to be variable specific.
• For example, BWSTRESU variable is populated by usage of only a single term “g”. However, in define.xml BWSTRESU variable has reference to a Codelist (UNIT) which consists of 4 unique terms. Such metadata is misleading and does not describe study data correctly
• Another example of this issue is a case of Flag variables like BWBLFL which are limited to usage of only single term “Y”. Submitted sample define.xml has a Codelist (NY) with two terms “N” and “Y” assigned to such variables. Term “N” is not applicable for Flag varaibles.
• Merging different codelists across many variables means actually missing codelists for these variables because such approach does not explain study specific data collection process
Tim Gartner we have seen this item come up a few times in our review of SEND datasets. I hope that it relates to the topic that we are investigating with the Phuse group.
“There are abbreviations in MAORRES (for example) such as LT and RT that are not defined in the define file or SDRG. These are defined in the study report on an Individual Gross and Microscopic Findings Explanation Page. Additionally, these carry over into CO where there can be a number with the abbreviation (1, RT) with no explanation regarding the number.” [9]
Anne Lindberg
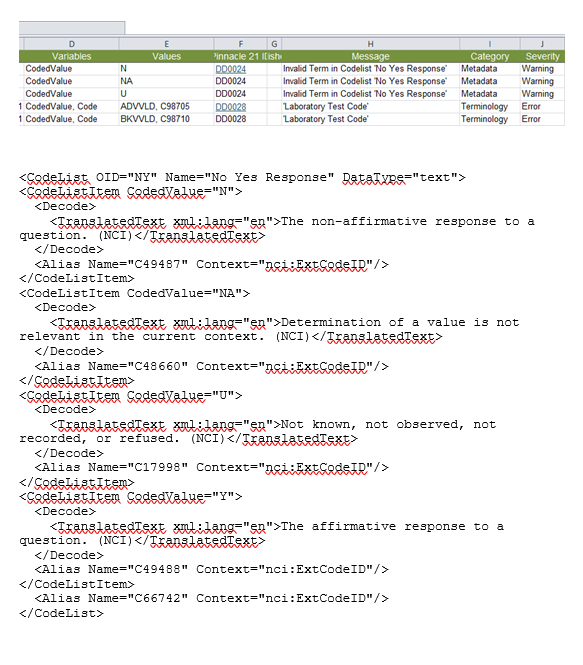
1. How are controlled terminologies with test name and test code presented correctly, which item types to use? (BGTEST & BGTESTCD, LBTEST & LBTESTCD etc) Define-xml specification, section 4.3.1.2 shows an example in which CT of test names is presented using EnumeratedItems and CT of test codes using CodeListItems. In some define files I have seen CodeListItems are used both for test name and test code. When to use Enumerated and when CodeLists? Does it depend on if the CT is extensible? Example from a define.xml: [6]
2. How does FDA use the CT of a define.xml [1]: is it important to include all available values from SEND terminology even if only some of them are used in the submitted SEND data set? [3]
Example: All SEND values from AGEU (age unit) are listed in define.xml even if only WEEKS is used in the data set. Especially the LBTEST list is growing and growing.
3. We have extended LBTEST with our custom terminology. When listing the code list in define.xml the custom values (extended values) actually used in the submitted data set must be present, but is it wrong to include custom values that were not present in this data set but are used in another study? That is, can we add the same set of extended values to every define.xml or do we need to customize it for each study [3]
4. We still receive many define.xml version 1 files. If possible, it would be nice to add a note to the final recommendations of this working group if a specific recommendation does not apply to define v1. Something like “this does not apply to define v1”. If most of the recommendations will be define v2 specific, then such notes are not needed.
Note from J. Feldmann: need clarification - who do you want to get this note from on the define v1 file? Anne: I tried to rephrase the question.
5. When checking the pinnacle validation report about a define.xml, I cannot always find where the issue is. Is there a list of know issues or help with troubleshooting. Example: A warning about an invalid term with category “metadata” and not “terminology” in Pinnacle validation report. Does this refer to an error in terminology section or somewhere else? [10]
Participant 8
1. Origin is always not very clear to define for the variables. IG described but it's still hard to follow. Can we clarify for it?
2. We got question about "When a variable/value has an Origin of Derived, it is best practice to populate the Comments with the algorithm used to derive the values." however, it cannot be for every variable. Now we defined all TEST or TESTCD as Derived variables, and we don't have algorithm for the CT mapping. What's the appropriate way to do so, change the Origin or Comment as mapping?
Note from J. Feldmann: not in scope for this project
Participant 9
1. For each of the SEND fields, what are the applicable reasons for providing code lists. The answer to this can help us know what to supply. Here are some possible answers for the purpose:
A. To see the decoded value to better understand the observation?
B. To see what was available during the study for selection, but was not observed on the study?
C. Other reasons?
[1]
2. When a computer system's data collection menu includes a sub-set of an external code list augmented by some additional terms, what is the best way to represent this in define? If purpose 1.A is all that is needed, can we supply a <CodeList> with both a <CodeListRef> and a <CodeListItem>? If purpose 1.B needs to be satisfied, then we presumably need to list each item using <CodeListItem> tags. Correct? [3?]
3.The use of <EnumeratedItem> tags doesn’t support purpose 1.A, but could satisfy purpose 1.B. Is this the only use for this tag? If not, in what other cases? [6]
Participant 10
What is the expectation of the level of inclusion of the ‘codelist’ in define.xml by the FDA reviewers. Currently, our define.xml automatically includes the entirety of the codelists used without any mapping; i.e. a regurgitation of the CDISC codelist version used in dataset production without any mapping. Is there an expectation of a) these codelists with ANY mapping done internally by the organization, b) the codelists (inclusive) with mapping of terms that are not obvious (e.g. GLUC = GLU is obvious, 1 of 5 = MILD is not obvious), c) Only terms from codelists used on the study, d) terms from the codelists used on the study WITH mapping, e) other? [3]
Participant 11
1) It is my understanding that only the terminology actually used in the SEND datasets should be included in the define file. How does this apply to scoring scales when not all terms in the scoring scale are used on study? Should the whole scoring scale be included in the define file? [3]
2) Where should scoring scales ideally be decoded: in the SEND dataset, in the nSDRG, in the define file, in all 3? [3]
Participant 12
We recently received feedback from a pilot SEND submission (see below). Most comments were around the define file and we are seeking to mitigate those. One of our mitigations was regarding codelists and we are wondering if the resolution we are proposing is a valid approach? (we had not received this comment on a previous test submission!)
Reviewer Comment: Some Codelists are merged across many variables
• Codelist should describe variable collection or derivation. Therefore, Codelist is expected to be variable specific.
• For example, BWSTRESU variable is populated by usage of only a single term “kg”. However, in define.xml BWSTRESU variable has reference to a Codelist (UNIT) which consists of 17 unique terms. Such metadata is misleading and does not describe study data correctly.
• Another example of this issue is a case of Flag variables like BWBLFL which are limited to usage of only single term “Y”. Submitted sample define.xml has a Codelist (NY) with two terms “N” and “Y” assigned to such variables. Term “N” is not applicable for Flag variables.
• Merging different codelists across many variables means actually missing codelists for these variables because such approach does not explain study specific data collection process.
We are therefore going to create new but specific code lists to address this. In one study we had age represented in Months and Days so previously AGU listed both units. We therefore differentiated this creating two new code lists ‘TS_AGEU’ (years) and DM_AGU (days). We are also now planning to apply this to other parameters e.g. create specific UNIT lists e.g. for the BW domain (Codelist BW_UNIT) where previously the UNIT code list listed all units for the study. Similarly this would apply to other domains where we want them to link to specific units entries e.g. LB domain and SPEC, DOSE and possibly SEV (for CLIN and MI/MA).
So adding (for example) the following codelists:
BW_UNIT
LB_UNIT
EXDOSEU_UNIT
Etc
These would have to be specific to each study but hopefully there would be a core set [4].
The other question was to do with dictionaries – We had a comment when we stated ‘no dictionaries used’ which was picked up “There is no such Codelist as “No Dictionaries Used” this is a comment not a codelist. This may be the way our define software works. We gathered from that that we should have no reference to dictionaries at all if we have not used any ‘external’ dictionaries so we removed it. But was this really necessary? [11]
J. Feldmann
1. When a variable entry contains more than one value (separated by a semi-colon) do you include two entries in the codelist? [3]
2. If there is only one choice possible, must it be in a codelist? For example, if Null and Y only, does Y have to be included in a codelist?
3. Which variables that are not associated with published CDISC controlled terminology must have a define.xml codelist? For example, ARM and SET have a define list of entries from the protocol, so should those have a codelist? [2]
4. When is it appropriate to include terms not used in the study (specific rules)? My thinking is that you do this when the full list is needed to interpret the entries that have been used on the study, for example when there is a ranked list of terms, you need to have all entries available for collection shown to correctly interpret what was collected on the study. [3]
5. When an "on the fly" entry is made during collection (i.e., an entry is made that is NOT from an available list), is that included in the codelist, since the codelist is the "set of allowable values"? Is an "on the fly" entry reported any differently than entries from a planned list? For example - clinical signs. [3]
6. How are different protocol-required tissue lists included in codelists? For example, if the required tissues for micro for controls is different form the required tissues for the high dose group. In this case, is there more than one codelist for the same test (micro)? There is no variable in the data that can break the entries up into value-level metadata for the codelist reference.
7. Do all values possible for pH need a codelist? [3]
8. Is there any specific naming convention that should be used for codelists that contains a subset of published controlled terminology? Should it always include the CDISC codelist name within the custom name (UNIT_LABS, UNIT_WEIGHT for example) or does it matter? [5]